Foraging toolkit demo - approximate inference analysis on Central Park birds data¶
Outline¶
Introduction¶
This notebook demonstrates using the foraging toolkit to analyze movements of birds recorded in Central Park, NYC, as described in the paper [1], Fig. 4.
The users are advised to read through the demo notebooks in `docs/foraging/random-hungry-followers <../random-hungry-followers/>`__ folder to familiarize themselves with the foraging toolkit, and, specifically, with computing predictors and running inference.
[1] R. Urbaniak, M. Xie, and E. Mackevicius, “Linking cognitive strategy, neural mechanism, and movement statistics in group foraging behaviors,” Sci Rep, vol. 14, no. 1, p. 21770, Sep. 2024, doi: 10.1038/s41598-024-71931-0.
[1]:
# importing packages. See https://github.com/BasisResearch/collab-creatures for repo setup
import logging
import os
import dill
import matplotlib.pyplot as plt
import pandas as pd
import collab.foraging.toolkit as ft
from collab.foraging.toolkit import dataObject, plot_predictor
logging.basicConfig(format="%(message)s", level=logging.INFO)
from collab.utils import find_repo_root
root = find_repo_root()
import plotly.io as pio
pio.renderers.default = "notebook"
# users can ignore smoke_test -- it's for automatic testing on GitHub,
# to make sure the notebook runs on future updates to the repository
smoke_test = "CI" in os.environ
frames = 150 if smoke_test else 300
frames_sps = 150 if smoke_test else 300
sampling_rate = 0.001 if smoke_test else 0.01
num_svi_iters = 100 if smoke_test else 1000
num_samples = 100 if smoke_test else 1000
Data processing¶
Load data¶
First, we convert the raw data into foraging toolkit format. Our methods expect a dataObject with foragersDF property, which is a DataFrame with columns time,forager,x,y. We use the toolkit utilities to subsample and rescale the data.
Notice that NaN values are valid, indicating that a certain forager was not detected in a certain time frame. The warning remains, as depending on the hypotheses and goals of a researcher, one might want to make a decision whether frames where at least one animal disappears are pathological and should be disposed of. In this case, we expect some missing data, as the birds were observed in the wild within a limited area, and sometimes leave the field of view.
[2]:
ducks_raw = pd.read_csv(
os.path.join(
root,
"data/foraging/central_park_birds_cleaned_2022/20221215122046189_-5_25_bone.avi.hand_checked_cleaned_df.csv",
)
)
sparrows_raw = pd.read_csv(
os.path.join(
root,
"data/foraging/central_park_birds_cleaned_2022/20221229124843603_n5_25_bone.avi.hand_checked_cleaned_df.csv",
)
)
ducks_raw = ducks_raw.rename(columns={"bird": "forager"})
sparrows_raw = sparrows_raw.rename(columns={"bird": "forager"})
ducks_sub = ft.subset_frames_evenly_spaced(ducks_raw, frames)
ducks_sub = ft.rescale_to_grid(ducks_sub, 90)
ducks_object = dataObject(foragersDF=ducks_sub)
sps_sub = ft.subset_frames_evenly_spaced(sparrows_raw, frames_sps)
sps_sub = ft.rescale_to_grid(sps_sub, 90)
sps_object = dataObject(foragersDF=sps_sub)
print(
f"\nDucks:\n------\n {ducks_object.foragersDF.head()}\n\nSparrows:\n--------\n {sps_object.foragersDF.head()}"
)
original_frames: 1451
original_shape: (8867, 4)
resulting_frames: 300
resulting_shape: (1848, 4)
original_frames: 11550
original_shape: (22821, 4)
resulting_frames: 279
resulting_shape: (617, 4)
Ducks:
------
x y time forager
0 NaN NaN 0 0
1 NaN NaN 1 0
2 NaN NaN 2 0
3 NaN NaN 3 0
4 NaN NaN 4 0
Sparrows:
--------
x y time forager
0 NaN NaN 0 0
1 NaN NaN 1 0
2 NaN NaN 2 0
3 NaN NaN 3 0
4 NaN NaN 4 0
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 0, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 1, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 2, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 3, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 4, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 5, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 6, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 7, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 8, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 9, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 10, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 11, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 12, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 13, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 14, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 15, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 16, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 17, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 18, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 19, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 20, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 21, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 22, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 23, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 24, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 25, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 26, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 27, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 28, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 0, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 1, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 2, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 3, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 4, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 5, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 6, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 7, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 8, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 9, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 10, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 11, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 12, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 13, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 14, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 15, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 16, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 17, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 18, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 19, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 20, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 21, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 22, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 23, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 24, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 25, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 26, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/utils.py:62: UserWarning:
Missing frames encountered for forager 27, adding NaN fillers.
The default behavior of predictor/score generating functions is
to ignore foragers with missing positional data. To modify, see documentation of
`derive_predictors_and_scores` and `generate_local_windows`
Visualize trajectories¶
[3]:
fig, ax = plt.subplots(1, 2, figsize=(12, 6))
ax1 = ft.plot_trajectories(ducks_raw, "ducks", ax=ax[0], show_legend=False)
ax2 = ft.plot_trajectories(sparrows_raw, "sparrows et al.", ax=ax[1], show_legend=False)
path = os.path.join(root, "docs/figures/fig_bird_trajectories.png")
if not os.path.exists(path):
plt.savefig(path, dpi=300)
plt.show()
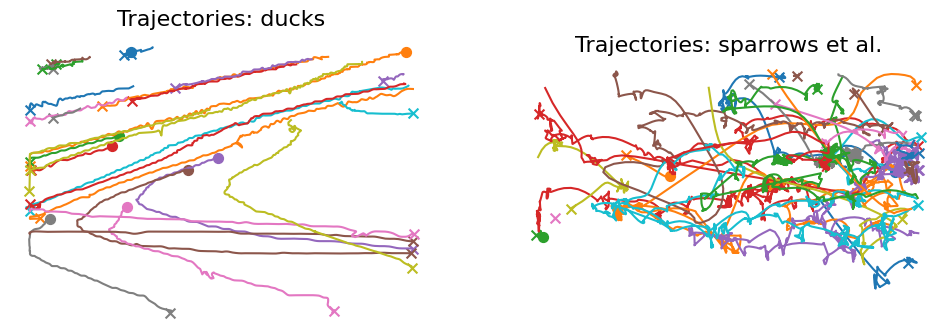
Analyze mutual distance distribution¶
The ft.foragers_to_forager_distances helper function calculates the distances between foragers at each time frame. It returns a nested list of length len(obj.foragers) where sublist i contains DataFrames of distances between forager i and all others, one DataFrame per time frame.
This takes a few minutes to run, and results are stored in local .pkl files, which you should delete if you want to re-compute the analysis.
[4]:
duck_distances_path = os.path.join(
root, "docs/foraging/central-park-birds/duck_distances.pkl"
)
sps_distances_path = os.path.join(
root, "docs/foraging/central-park-birds/sps_distances.pkl"
)
if not os.path.exists(duck_distances_path) or not os.path.exists(sps_distances_path):
duck_distances = ft.foragers_to_forager_distances(obj=ducks_object)
sps_distances = ft.foragers_to_forager_distances(obj=sps_object)
with open(duck_distances_path, "wb") as f:
dill.dump(duck_distances, f)
with open(sps_distances_path, "wb") as f:
dill.dump(sps_distances, f)
else:
print("Loading precomputed distances.")
with open(duck_distances_path, "rb") as f:
duck_distances = dill.load(f)
with open(sps_distances_path, "rb") as f:
sps_distances = dill.load(f)
Loading precomputed distances.
Here we are using plot_distances to conveniently plot the distance histogram over the entire simulation. See Fig. 4(E,F) in [1].
[5]:
fig = ft.plot_distances(duck_distances, "ducks")
fig.show()
path = os.path.join(root, "docs/figures/fig_distance_distribution_ducks.png")
if not os.path.exists(path):
fig.write_image(path, scale=2)
fig = ft.plot_distances(sps_distances, "sparrows et al.")
fig.show()
path = os.path.join(root, "docs/figures/fig_distance_distribution_sps.png")
if not os.path.exists(path):
fig.write_image(path, scale=2)
Inference with a specific quantitative hypothesis¶
The foraging toolkit can be used to formulate certain parametrized hypotheses regarding certain behaviors, fit different models to the data and compare the strength of the fitted coefficients. Assuming reasonable normalization, stronger coefficients correspond to hypotheses that are more consistent with the data. An example of such analysis is given in [1], Fig. 4(G,H), where the interaction_length parameter in proximity predictor was varied.
Here we provide an example for computing a single fit (similar to the other demo notebooks), with a hand-selected preferred proximity (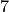 ) which is expected to be a better fit for ducks than for sparrows.
) which is expected to be a better fit for ducks than for sparrows.
In the future, in a separate notebook, we will illustrate a systematic approach to hypothesis choice.
Predictors derivation¶
Compute the relevant derived predictors (proximity and access), which will be used later for inference.
[6]:
def derive_cp_predictors(data_object, rep, opt):
window_size = 30
sampling_fraction = 0.2
score_kwargs = {
"nextStep_sublinear_25": {
"nonlinearity_exponent": 2.5
}, # chosen to decrease nonlinearity in I/O relationship
}
predictor_kwargs = {
"access": {
"decay_factor": 0.9,
},
"proximity": {
"interaction_length": 90,
"optimal_distance": opt,
"repulsion_radius": rep,
"proximity_decay": 0.2,
},
}
local_windows_kwargs = {
"window_size": window_size,
"sampling_fraction": sampling_fraction,
"skip_incomplete_frames": False,
}
object_derived_DF = ft.derive_predictors_and_scores(
data_object,
local_windows_kwargs,
predictor_kwargs=predictor_kwargs,
score_kwargs=score_kwargs,
dropna=True,
add_scaled_values=True,
)
return object_derived_DF
# we will use the same parameters for both species
ducks_derivedDF = derive_cp_predictors(ducks_object, rep=2, opt=7)
sps_derivedDF = derive_cp_predictors(sps_object, rep=2, opt=7)
2024-10-28 16:49:11,062 - access completed in 1.27 seconds.
2024-10-28 16:49:21,107 - proximity completed in 10.04 seconds.
2024-10-28 16:49:22,060 - nextStep_sublinear_25 completed in 0.95 seconds.
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/derive.py:56: UserWarning:
Dropped 19072/1705366 frames from `derivedDF` due to NaN values.
Missing values can arise when computations depend on next/previous step positions
that are unavailable. See documentation of the corresponding predictor/score generating
functions for more information.
2024-10-28 16:49:25,006 - access completed in 0.88 seconds.
2024-10-28 16:49:26,372 - proximity completed in 1.37 seconds.
2024-10-28 16:49:26,687 - nextStep_sublinear_25 completed in 0.31 seconds.
/Users/emily/code/collaborative-intelligence/collab/foraging/toolkit/derive.py:56: UserWarning:
Dropped 22266/621519 frames from `derivedDF` due to NaN values.
Missing values can arise when computations depend on next/previous step positions
that are unavailable. See documentation of the corresponding predictor/score generating
functions for more information.
Predictors visualization¶
Plot the proximity score for a particular forager (forager_predictor_indices=[2]) at different timesteps.
[7]:
predictor_name = "proximity"
plot_predictor(
ducks_object.foragers,
ducks_object.derived_quantities[predictor_name],
predictor_name=predictor_name,
time=range(47, 55),
grid_size=90,
size_multiplier=10,
random_state=99,
forager_position_indices=list(range(29)),
forager_predictor_indices=[2],
)
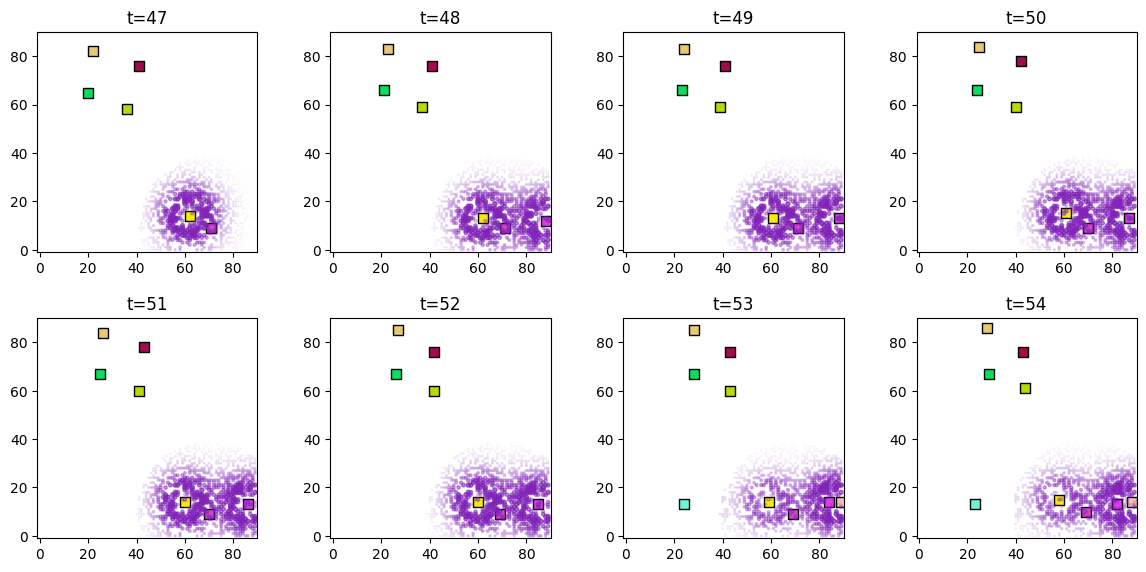
[8]:
predictor_name = "proximity"
plot_predictor(
sps_object.foragers,
sps_object.derived_quantities[predictor_name],
predictor_name=predictor_name,
time=range(9, 12),
grid_size=90,
size_multiplier=10,
random_state=99,
forager_position_indices=list(range(28)),
forager_predictor_indices=[2],
)
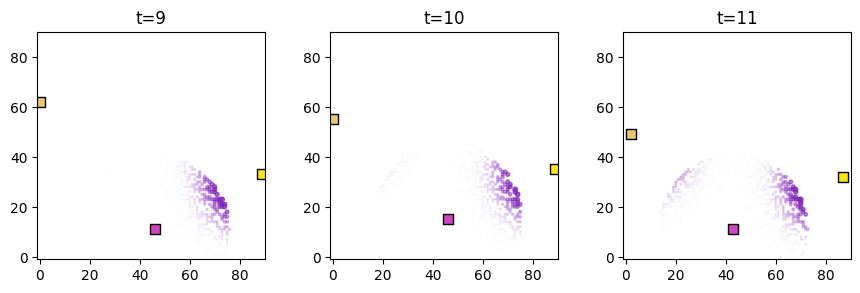
Model fit¶
Estimate model coefficients using SVI.
[9]:
predictors = ["access", "proximity"]
outcome_vars = ["nextStep_sublinear_25"]
ducks_predictor_dict, ducks_outcome_dict = ft.prep_DF_data_for_inference(
ducks_derivedDF,
predictors,
outcome_vars,
subsample_rate=0.1,
)
ducks_model = ft.HeteroskedasticLinear(ducks_predictor_dict, ducks_outcome_dict)
ducks_outcome = ft.get_samples(
model=ducks_model,
predictors=ducks_predictor_dict,
outcome=ducks_outcome_dict,
num_svi_iters=num_svi_iters,
num_samples=num_samples,
plot=False,
verbose=False,
)
sps_predictor_dict, sps_outcome_dict = ft.prep_DF_data_for_inference(
sps_derivedDF,
predictors,
outcome_vars,
subsample_rate=0.1,
)
sps_model = ft.HeteroskedasticLinear(sps_predictor_dict, sps_outcome_dict)
sps_outcome = ft.get_samples(
model=sps_model,
predictors=sps_predictor_dict,
outcome=sps_outcome_dict,
num_svi_iters=num_svi_iters,
num_samples=num_samples,
plot=False,
verbose=False,
)
2024-10-28 16:49:28,088 - Sample size: 168629
2024-10-28 16:49:28,089 - Starting SVI inference with 1000 iterations.
[iteration 0200] loss: 340825.9375
[iteration 0400] loss: 340222.9688
[iteration 0600] loss: 340346.9375
[iteration 0800] loss: 340113.4688
2024-10-28 16:49:38,251 - SVI inference completed in 10.16 seconds.
[iteration 1000] loss: 340413.5625
2024-10-28 16:49:39,011 - Sample size: 59925
2024-10-28 16:49:39,012 - Starting SVI inference with 1000 iterations.
[iteration 0200] loss: 116830.9219
[iteration 0400] loss: 116779.1016
[iteration 0600] loss: 116588.3906
[iteration 0800] loss: 116632.0000
2024-10-28 16:49:45,801 - SVI inference completed in 6.79 seconds.
[iteration 1000] loss: 116547.4297
Results¶
Finally, we can compare the fit quality. This can be done utilizing evaluate_performance method in the foraging toolkit.
Ducks¶
[10]:
ft.evaluate_performance(
model=ducks_model,
guide=ducks_outcome["guide"],
predictors=ducks_predictor_dict,
outcome=ducks_outcome_dict,
num_samples=1000,
)
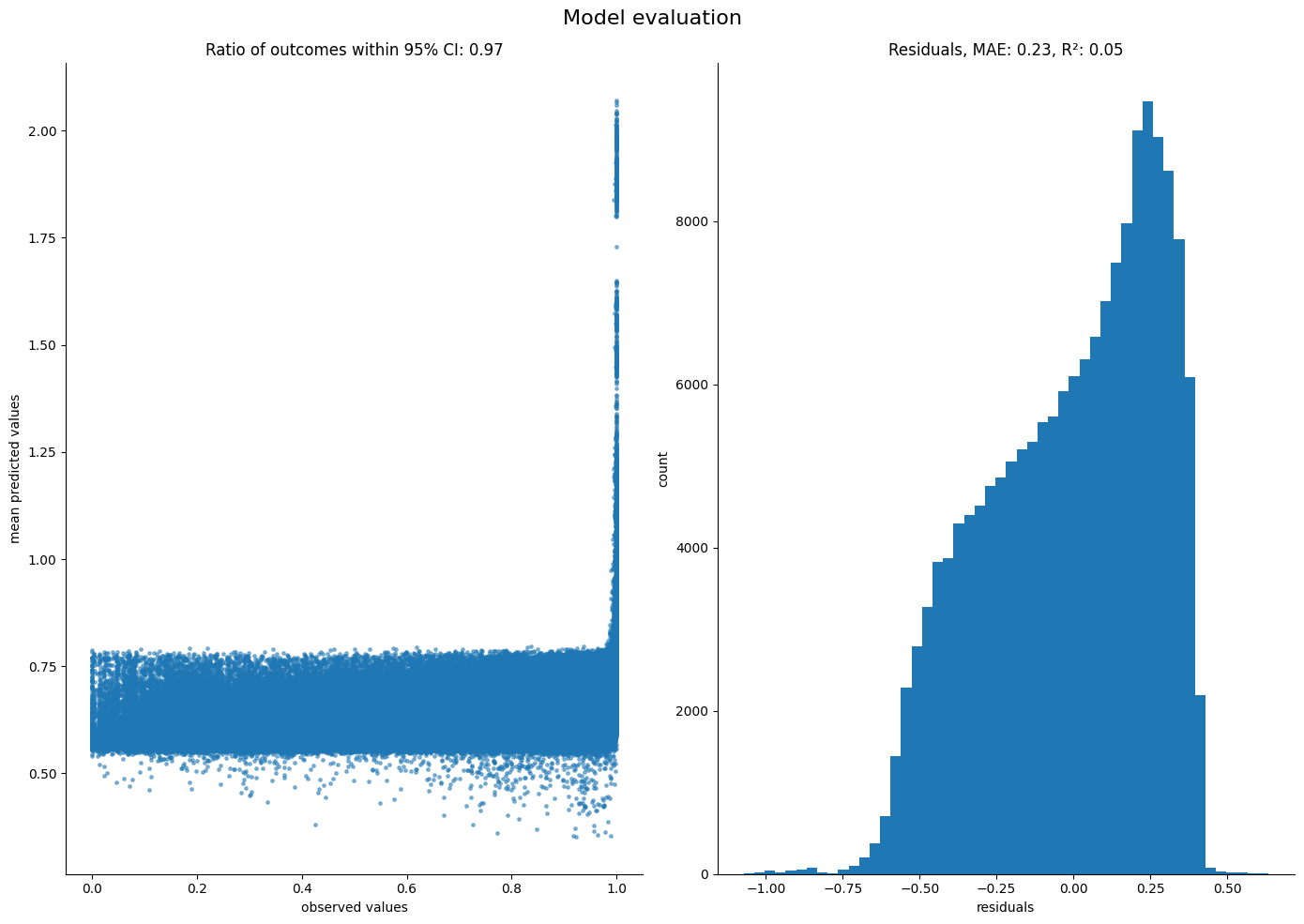
Poor 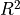 fit suggests that while the model explains some variance, clearly there are many other things that guide the birds that this model does not capture.
fit suggests that while the model explains some variance, clearly there are many other things that guide the birds that this model does not capture.
An interesting direction will be to tweak the score functional form.
Sparrows¶
[11]:
ft.evaluate_performance(
model=sps_model,
guide=sps_outcome["guide"],
predictors=sps_predictor_dict,
outcome=sps_outcome_dict,
num_samples=1000,
)
The model provides an even poorer fit to the data.
Comparison¶
Now let us compare the fit coefficients for the proximity predictor.
[12]:
new_key = f"weight_continuous_proximity_{outcome_vars[0]}"
sites = {
"ducks": ducks_outcome["samples"][new_key],
"sparrows et al.": sps_outcome["samples"][new_key],
}
ft.plot_coefs(
sites,
"Posterior marginal for the proximity coefficients<br>(both models)",
nbins=120,
ann_start_y=160,
ann_break_y=50,
)
Interestingly, negative coefficients suggest that actually the sparrows prefer to avoid large separations.