Foraging toolkit demo - communicating foragers, Part II (Inference)¶
Outline¶
Introduction¶
In a multi-agent context, communication of information between foragers is an important feature of group-level behavior, as is the impact of different environmental conditions. We thus explore how one might infer to what extent agents communicate with each other to facilitate foraging. We ask whether the benefit of communicating information would be different for different environments, using multiple simulations with a range of communication-related hyper-parameters. In environments where food is highly clustered, it takes longer for birds to find food, but in all environments, using information communicated from other birds improves foraging success. The Bayesian inference methods are able to correctly compare the extent to which simulated agents communicate about the locations of the rewards.
The communicating foragers demo is divided into two notebooks:
Simulation
`communicators_simulations.ipynb<./communicators_simulations.ipynb>`__ - run/read this prior to continuing furtherInference (this one)
The users are advised to read through the demo notebooks in `docs/foraging/random-hungry-followers <../random-hungry-followers/>`__ folder to familiarize themselves with the foraging toolkit, and, specifically, computing predictors and running inference, which is the focus of this part of the demo.
The main reference is [1], in particular Fig.3.
[1] R. Urbaniak, M. Xie, and E. Mackevicius, “Linking cognitive strategy, neural mechanism, and movement statistics in group foraging behaviors,” Sci Rep, vol. 14, no. 1, p. 21770, Sep. 2024, doi: 10.1038/s41598-024-71931-0.
[1]:
# importing packages. See https://github.com/BasisResearch/collab-creatures for repo setup
import logging
import os
import time
import dill
import matplotlib.pyplot as plt
import pandas as pd
import plotly.io as pio
import collab.foraging.toolkit as ft
from collab.foraging.toolkit import dataObject
from collab.utils import find_repo_root
pio.renderers.default = "notebook"
root = find_repo_root()
logging.basicConfig(format="%(message)s", level=logging.INFO)
# users can ignore smoke_test -- it's for automatic testing on GitHub, to make sure the notebook runs on future updates to the repository
smoke_test = "CI" in os.environ
# smoke_test = True
num_frames = 5 if smoke_test else 50
num_svi_iters = 10 if smoke_test else 1000
num_samples = 10 if smoke_test else 1000
notebook_starts = time.time()
Load simulation data¶
We will analyze the two runs in the communicators_strong folder, corresponding to c_trust=0 (suffix  below) and
below) and c_trust=0.6 (suffix 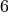 below). After loading the simulated data, we further make some data wrangling to create the foraging toolkit underlying data structure of type
below). After loading the simulated data, we further make some data wrangling to create the foraging toolkit underlying data structure of type ft.dataObject, which is further passed to the predictor computation routines. We also compute the local windows (see random_foragers.ipynb demo
notebook). We save the results on disk to speed up further experimentation.
[2]:
sim_params = pd.read_csv(
os.path.join(
root, "data/foraging/communicators/communicators_strong/metadataDF.csv"
),
index_col=0,
)
sim_params.head()
[2]:
| c_trust | sight_radius | reward_patch_dim | sim index | |
|---|---|---|---|---|
| 0 | 0.0 | 5 | 4 | 0 |
| 1 | 0.6 | 5 | 4 | 1 |
[3]:
always_generate = True
communicators_object_path0 = "communicators0.pkl"
communicators_object_path6 = "communicators6.pkl"
local_windows_kwargs = {
"window_size": 10,
"sampling_fraction": 1,
"skip_incomplete_frames": False,
}
if (not os.path.exists(communicators_object_path0)) or always_generate:
home_dir = os.path.join(root, "data/foraging/communicators/communicators_strong")
sim0_folder = "sim0_run0"
sim6_folder = "sim1_run0"
sim0_dir = os.path.join(home_dir, sim0_folder)
sim6_dir = os.path.join(home_dir, sim6_folder)
bird0 = pd.read_csv(os.path.join(sim0_dir, "foragerlocsDF.csv"), index_col=0)
foragerlocsDF0 = pd.read_csv(
os.path.join(sim0_dir, "foragerlocsDF.csv"), index_col=0
)
foragerlocsDF6 = pd.read_csv(
os.path.join(sim6_dir, "foragerlocsDF.csv"), index_col=0
)
rewardlocsDF0 = pd.read_csv(os.path.join(sim0_dir, "rewardlocsDF.csv"), index_col=0)
rewardlocsDF6 = pd.read_csv(os.path.join(sim6_dir, "rewardlocsDF.csv"), index_col=0)
# drop last frame to make the two dataframes the same length
last = foragerlocsDF0["time"].unique()[-1]
foragerlocsDF0 = foragerlocsDF0.drop(
foragerlocsDF0[foragerlocsDF0["time"] == last].index
)
foragerlocsDF6 = foragerlocsDF6.drop(
foragerlocsDF6[foragerlocsDF6["time"] == last].index
)
assert all(rewardlocsDF0["time"].unique() == foragerlocsDF0["time"].unique())
assert all(rewardlocsDF6["time"].unique() == foragerlocsDF6["time"].unique())
def shift_to_start_with_0s(df):
df["time"] = df["time"] - df["time"].min()
df["forager"] = df["forager"] - df["forager"].min()
return df
foragerlocsDF0 = shift_to_start_with_0s(foragerlocsDF0)
foragerlocsDF6 = shift_to_start_with_0s(foragerlocsDF6)
rewardlocsDF0["time"] = rewardlocsDF0["time"] - 1
rewardlocsDF6["time"] = rewardlocsDF6["time"] - 1
communicators0 = dataObject(
foragersDF=foragerlocsDF0, rewardsDF=rewardlocsDF0, grid_size=35
)
communicators6 = dataObject(
foragersDF=foragerlocsDF6, rewardsDF=rewardlocsDF6, grid_size=35
)
communicators0.foragersDF.head(), communicators6.foragersDF.head()
for foragers_object in [communicators0, communicators6]:
foragers_object.local_windows_kwargs = local_windows_kwargs
foragers_object.local_windows = ft.generate_local_windows(foragers_object)
with open(communicators_object_path0, "wb") as f:
dill.dump(communicators0, f)
with open(communicators_object_path6, "wb") as f:
dill.dump(communicators6, f)
else:
print("Loading communicators objects from disk...")
with open(communicators_object_path0, "rb") as f:
communicators0 = dill.load(f)
with open(communicators_object_path6, "rb") as f:
communicators6 = dill.load(f)
assert communicators0.grid_size == communicators6.grid_size
Derived quantities¶
Please consult the corresponding section in the ``random_foragers.ipynb` notebook <../random-hungry-followers/random_foragers.ipynb#derived-quantities>`__ for explanation of derived quantities (predictors and scores).
Here in addition to the three predictors described there, we will use a new communication predictor.
[4]:
# lists all the predictors available in the toolkit
ft.get_list_of_predictors()
[4]:
['access', 'communication', 'food', 'pairwiseCopying', 'proximity', 'vicsek']
[5]:
help(ft.generate_communication_predictor)
Help on function generate_communication_predictor in module collab.foraging.toolkit.communication:
generate_communication_predictor(foragers_object: collab.foraging.toolkit.utils.dataObject, predictor_name: str)
Generates communication-based predictors for a group of foragers. When a forager
is in the vicinity of food, it can communicate this information with the other
foragers. The predictor value is proportional to the proximity of the communicating
partner, but only if that partner is close to a food source.
The predictor can be customized by providing a custom communication function
(default: exponential decay) and/or a custom interaction function (default: closeness to food).
Arguments:
:param foragers_object: A data object containing information about the foragers, including their positions,
trajectories, and local windows. Such objects can be generated using `object_from_data`.
:param predictor_name: The name of the food predictor to be generated, used to fetch relevant parameters
from `foragers_object.predictor_kwargs` and to store the computed values.
:return: A list of lists of pandas DataFrames where each DataFrame has been updated with the computed food
predictor values.
Predictor-specific keyword arguments:
:param interaction_length: The maximum distance to the communicating partner.
:param interaction_constraint: An optional callable that imposes additional constraints on which
foragers can interact based on custom logic. Default is
`constraint_filter_close_to_reward`
:param interaction_constraint_params: Optional parameters to pass to the `interaction_constraint`
function. For `constraint_filter_close_to_reward`, this
includes `finders_tolerance` - the maximal distance
of the communicating partner to the food source.
:param communication_contribution_function: The decay function for computing the strength of the communication.
The value of the communication predictor will be equal to the total contribution from the
individual communicating partners.
The default value is the exponential decay function: f(dist) = exp(-decay_factor * dist).
The default decay factor is 0.5, it can be customized by passing
an additional `decay_factor` keyword argument.
In other words we have
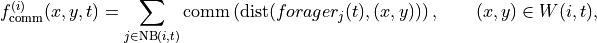
where
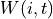 is the local window,
is the local window, is the communication function (default:
is the communication function (default: 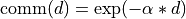 ), and
), and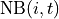 is the set of foragers interacting with
is the set of foragers interacting with 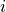 at time
at time 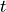 , where in addition, there exists
, where in addition, there exists  such that
such that 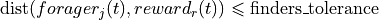 .
.
Compare this with the 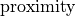 predictor in
predictor in `random_foragers.ipynb <../random-hungry-followers/random_foragers.ipynb#computation>`__.
The other predictors and the 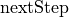 score are described in detail in
score are described in detail in `random_foragers.ipynb <../random-hungry-followers/random_foragers.ipynb>`__ as well.
Here we compute the derived quantities for the two simulations.
[6]:
communicators_derived_path0 = "communicators_derivedDF_0.pkl"
communicators_derived_path6 = "communicators_derivedDF_6.pkl"
always_generate = True
interaction_length = 30
if always_generate or smoke_test or not os.path.exists(communicators_derived_path0):
visibility_range = 6
interaction_constraint_params = {
"finders_tolerance": 3,
}
predictor_kwargs = {
"food": {
"interaction_length": 6, # as in simulations food placed further plays no role
"decay_factor": 0.5,
},
"communication": {
"interaction_length": interaction_length,
"interaction_minimal_distance": visibility_range + 1,
"interaction_constraint_params": interaction_constraint_params,
"decay_factor": 0.15,
},
"access": {
"decay_factor": 0.15,
},
"proximity": {
"interaction_length": interaction_length,
"interaction_constraint": None,
"interaction_constraint_params": None,
"repulsion_radius": 4,
"optimal_distance": 8,
"proximity_decay": 1,
},
}
score_kwargs = {
"nextStep_linear": {"nonlinearity_exponent": 1},
"nextStep_sublinear": {"nonlinearity_exponent": 0.5},
}
derivedDF_0 = ft.derive_predictors_and_scores(
communicators0,
local_windows_kwargs,
predictor_kwargs=predictor_kwargs,
score_kwargs=score_kwargs,
dropna=True,
add_scaled_values=True,
)
derivedDF_6 = ft.derive_predictors_and_scores(
communicators6,
local_windows_kwargs,
predictor_kwargs=predictor_kwargs,
score_kwargs=score_kwargs,
dropna=True,
add_scaled_values=True,
)
if not smoke_test:
with open(communicators_derived_path0, "wb") as f:
dill.dump(derivedDF_0, f)
with open(communicators_derived_path6, "wb") as f:
dill.dump(derivedDF_6, f)
# the objects themselves have been updated, need to save them
with open(communicators_object_path0, "wb") as f:
dill.dump(communicators0, f)
with open(communicators_object_path6, "wb") as f:
dill.dump(communicators6, f)
else:
print("Loading derivedDF from disk...")
with open(communicators_derived_path0, "rb") as f:
derivedDF_0 = dill.load(f)
with open(communicators_derived_path6, "rb") as f:
derivedDF_6 = dill.load(f)
display(derivedDF_0.head())
display(derivedDF_6.head())
2024-10-31 15:11:32,055: food completed in 1.84 seconds.
2024-10-31 15:11:33,658: communication completed in 1.60 seconds.
2024-10-31 15:11:34,047: access completed in 0.39 seconds.
2024-10-31 15:11:36,957: proximity completed in 2.91 seconds.
2024-10-31 15:11:37,239: nextStep_linear completed in 0.28 seconds.
2024-10-31 15:11:37,513: nextStep_sublinear completed in 0.27 seconds.
/home/rafal/s78projects/collab-creatures/collab/foraging/toolkit/derive.py:56: UserWarning:
Dropped 2276/110448 frames from `derivedDF` due to NaN values.
Missing values can arise when computations depend on next/previous step positions
that are unavailable. See documentation of the corresponding predictor/score generating
functions for more information.
2024-10-31 15:11:40,618: food completed in 1.85 seconds.
2024-10-31 15:11:41,698: communication completed in 1.08 seconds.
2024-10-31 15:11:42,108: access completed in 0.41 seconds.
2024-10-31 15:11:44,367: proximity completed in 2.26 seconds.
2024-10-31 15:11:44,782: nextStep_linear completed in 0.41 seconds.
2024-10-31 15:11:45,076: nextStep_sublinear completed in 0.29 seconds.
/home/rafal/s78projects/collab-creatures/collab/foraging/toolkit/derive.py:56: UserWarning:
Dropped 2185/119521 frames from `derivedDF` due to NaN values.
Missing values can arise when computations depend on next/previous step positions
that are unavailable. See documentation of the corresponding predictor/score generating
functions for more information.
| x | y | distance_to_forager | time | forager | food | communication | access | proximity | distance_to_next_step | nextStep_linear | nextStep_sublinear | food_scaled | communication_scaled | access_scaled | proximity_scaled | nextStep_linear_scaled | nextStep_sublinear_scaled | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 13 | 20 | 10.000000 | 0 | 0 | 0.0 | 0.0 | 0.223130 | 0.851696 | 8.062258 | 0.339514 | 0.187297 | 0.0 | 0.0 | 0.000000 | 0.918505 | 0.339514 | 0.187297 |
| 1 | 14 | 16 | 9.848858 | 0 | 0 | 0.0 | 0.0 | 0.228247 | 1.000000 | 7.615773 | 0.376092 | 0.210121 | 0.0 | 0.0 | 0.006586 | 1.000000 | 0.376092 | 0.210121 |
| 2 | 14 | 17 | 9.486833 | 0 | 0 | 0.0 | 0.0 | 0.240984 | 0.911436 | 7.280110 | 0.403590 | 0.227724 | 0.0 | 0.0 | 0.022982 | 0.951333 | 0.403590 | 0.227724 |
| 3 | 14 | 18 | 9.219544 | 0 | 0 | 0.0 | 0.0 | 0.250842 | 0.837656 | 7.071068 | 0.420716 | 0.238893 | 0.0 | 0.0 | 0.035671 | 0.910790 | 0.420716 | 0.238893 |
| 4 | 14 | 19 | 9.055385 | 0 | 0 | 0.0 | 0.0 | 0.257095 | 0.835199 | 7.000000 | 0.426538 | 0.242727 | 0.0 | 0.0 | 0.043721 | 0.909439 | 0.426538 | 0.242727 |
| x | y | distance_to_forager | time | forager | food | communication | access | proximity | distance_to_next_step | nextStep_linear | nextStep_sublinear | food_scaled | communication_scaled | access_scaled | proximity_scaled | nextStep_linear_scaled | nextStep_sublinear_scaled | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 2 | 10.000000 | 0 | 0 | 0.0 | 0.0 | 0.223130 | 0.000718 | 10.816654 | 0.016668 | 0.008369 | 0.0 | 0.0 | 0.000000 | 0.500359 | 0.016668 | 0.008369 |
| 1 | 0 | 3 | 9.219544 | 0 | 0 | 0.0 | 0.0 | 0.250842 | 0.000983 | 10.000000 | 0.090909 | 0.046537 | 0.0 | 0.0 | 0.035671 | 0.500491 | 0.090909 | 0.046537 |
| 2 | 0 | 4 | 8.485281 | 0 | 0 | 0.0 | 0.0 | 0.280049 | 0.001392 | 9.219544 | 0.161860 | 0.084500 | 0.0 | 0.0 | 0.073266 | 0.500696 | 0.161860 | 0.084500 |
| 3 | 0 | 5 | 7.810250 | 0 | 0 | 0.0 | 0.0 | 0.309890 | 0.002203 | 8.485281 | 0.228611 | 0.121712 | 0.0 | 0.0 | 0.111679 | 0.501102 | 0.228611 | 0.121712 |
| 4 | 0 | 6 | 7.211103 | 0 | 0 | 0.0 | 0.0 | 0.339030 | 0.004166 | 7.810250 | 0.289977 | 0.157372 | 0.0 | 0.0 | 0.149189 | 0.502083 | 0.289977 | 0.157372 |
Visualization¶
[7]:
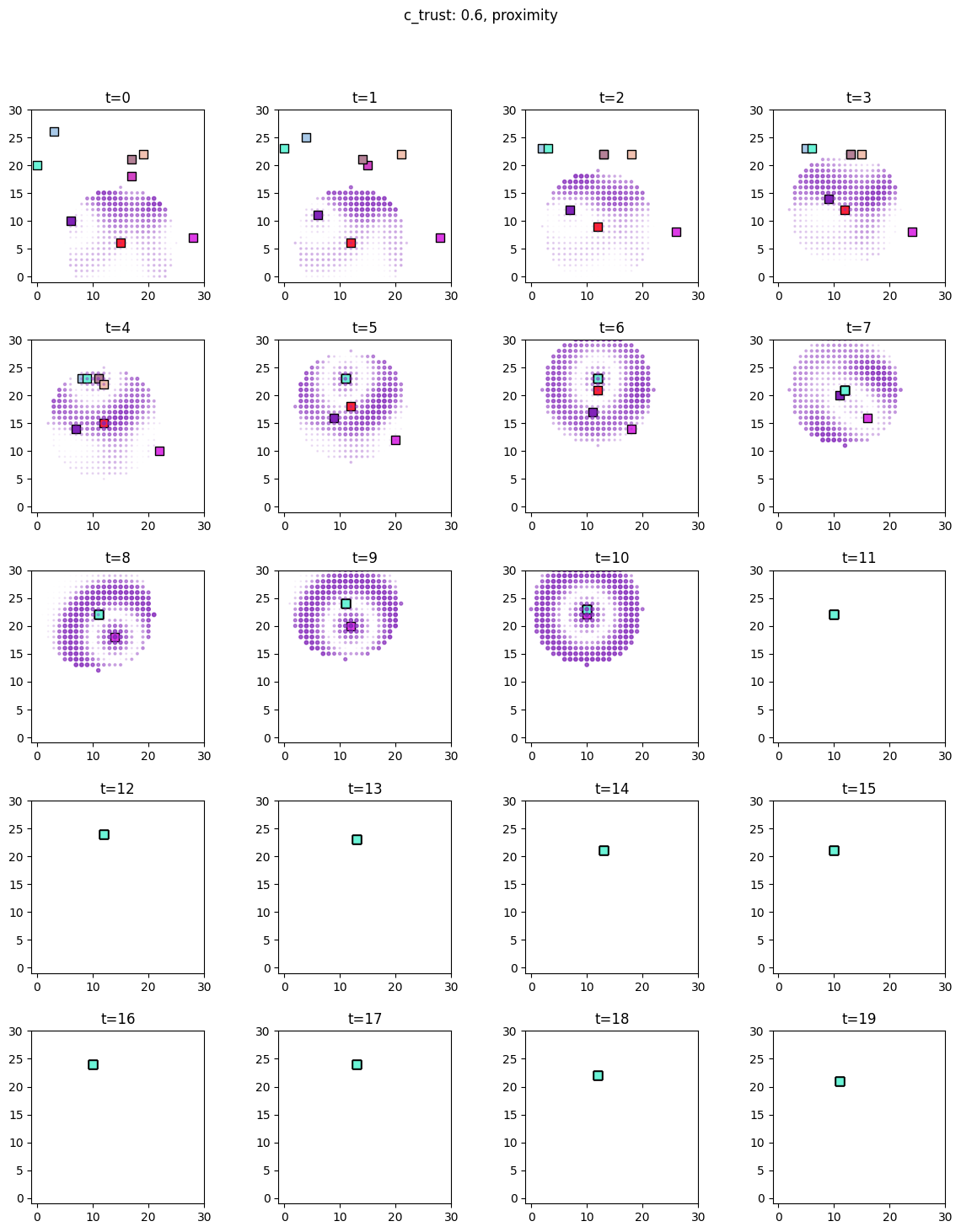
for i, sim in enumerate([communicators0, communicators6]):
for derived_quantity_name in [
"food",
"communication",
"proximity",
]: # sim.derived_quantities.keys():
ft.plot_predictor(
sim.foragers,
sim.derived_quantities[derived_quantity_name],
predictor_name=derived_quantity_name,
time=range(20),
grid_size=30,
size_multiplier=10,
random_state=99,
forager_position_indices=list(range(8)),
forager_predictor_indices=[4],
)
# pad suptitle 2mm
plt.subplots_adjust(top=0.9)
plt.suptitle(f"c_trust: {sim_params.iloc[i].c_trust}, {derived_quantity_name}")
plt.show()
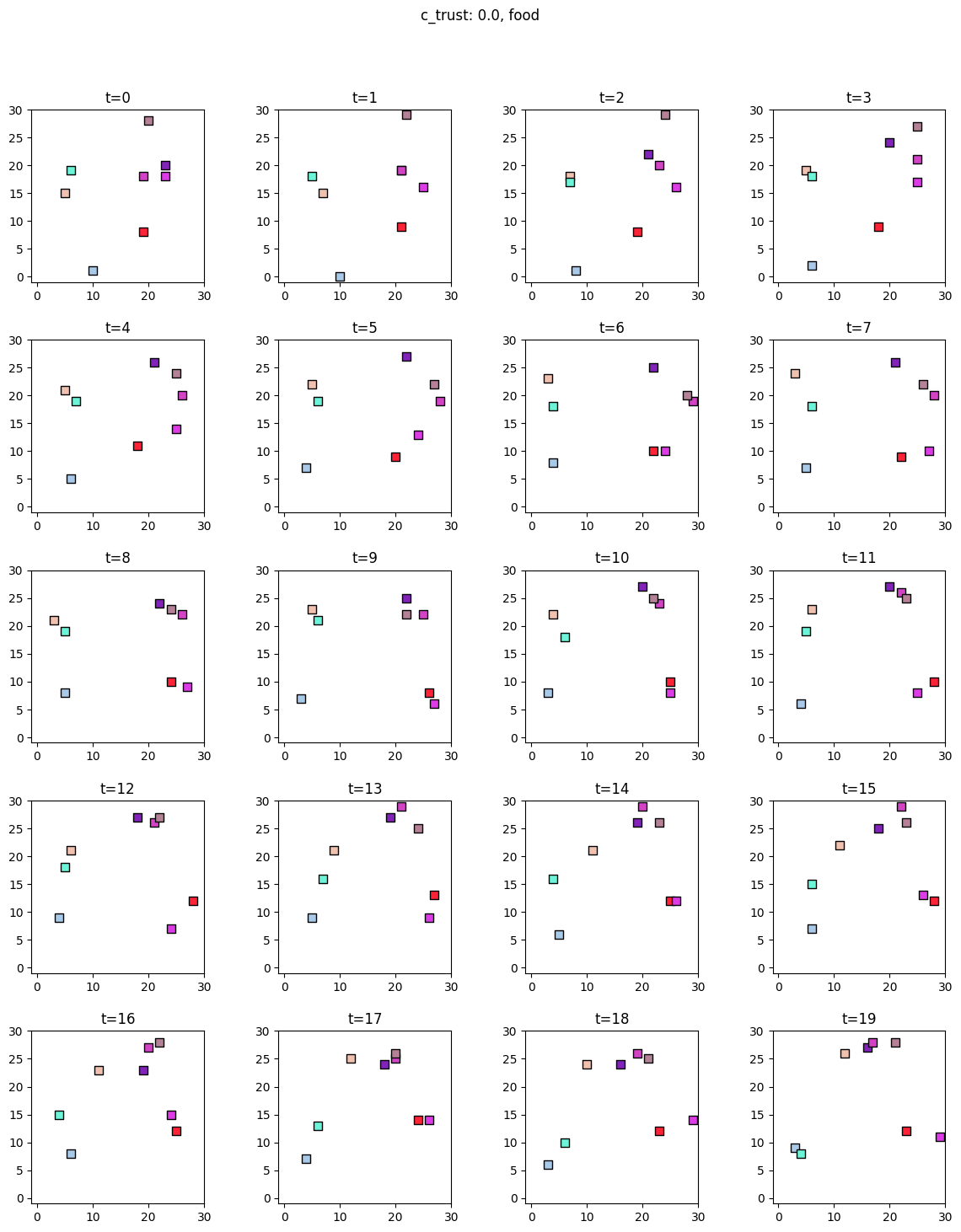
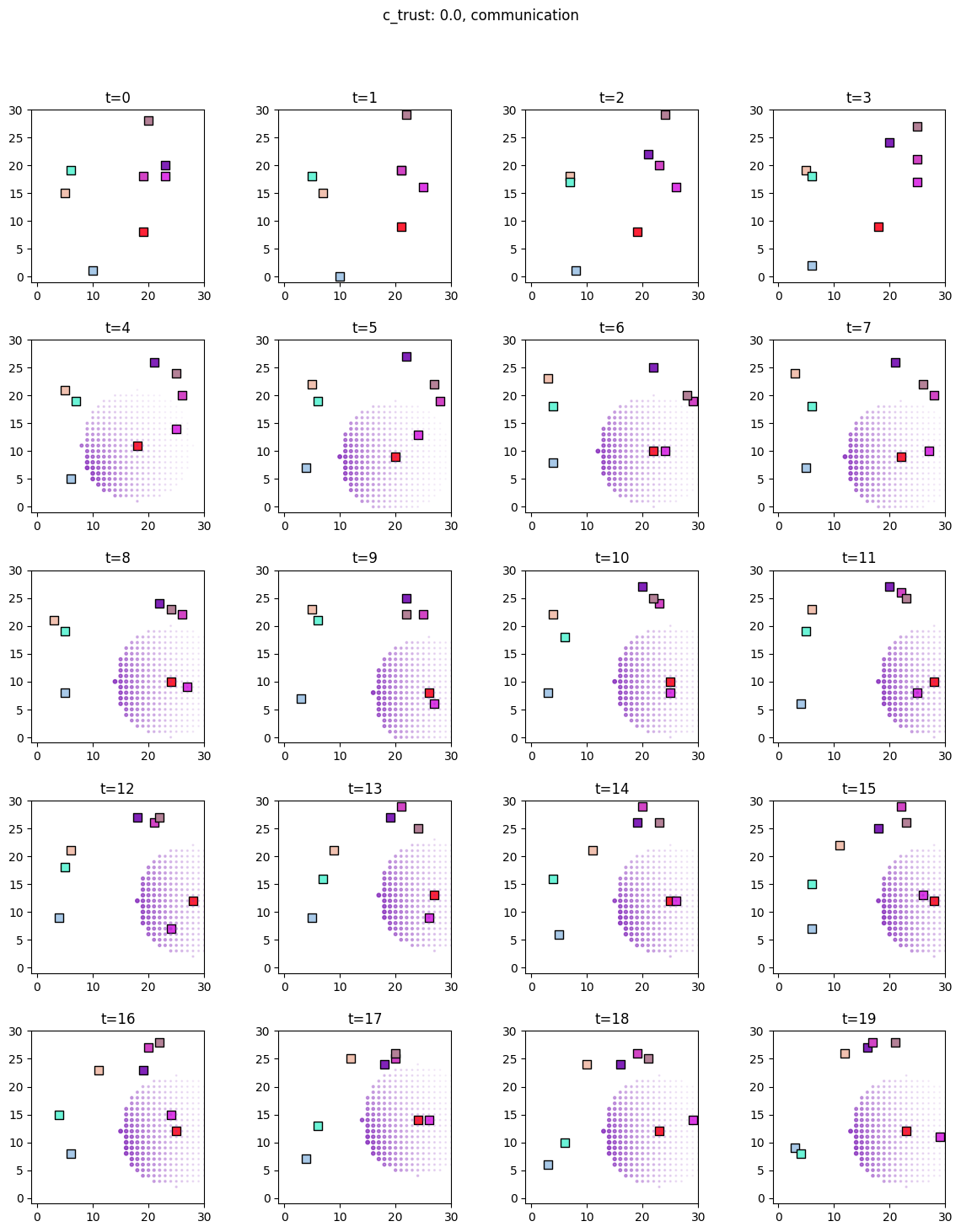
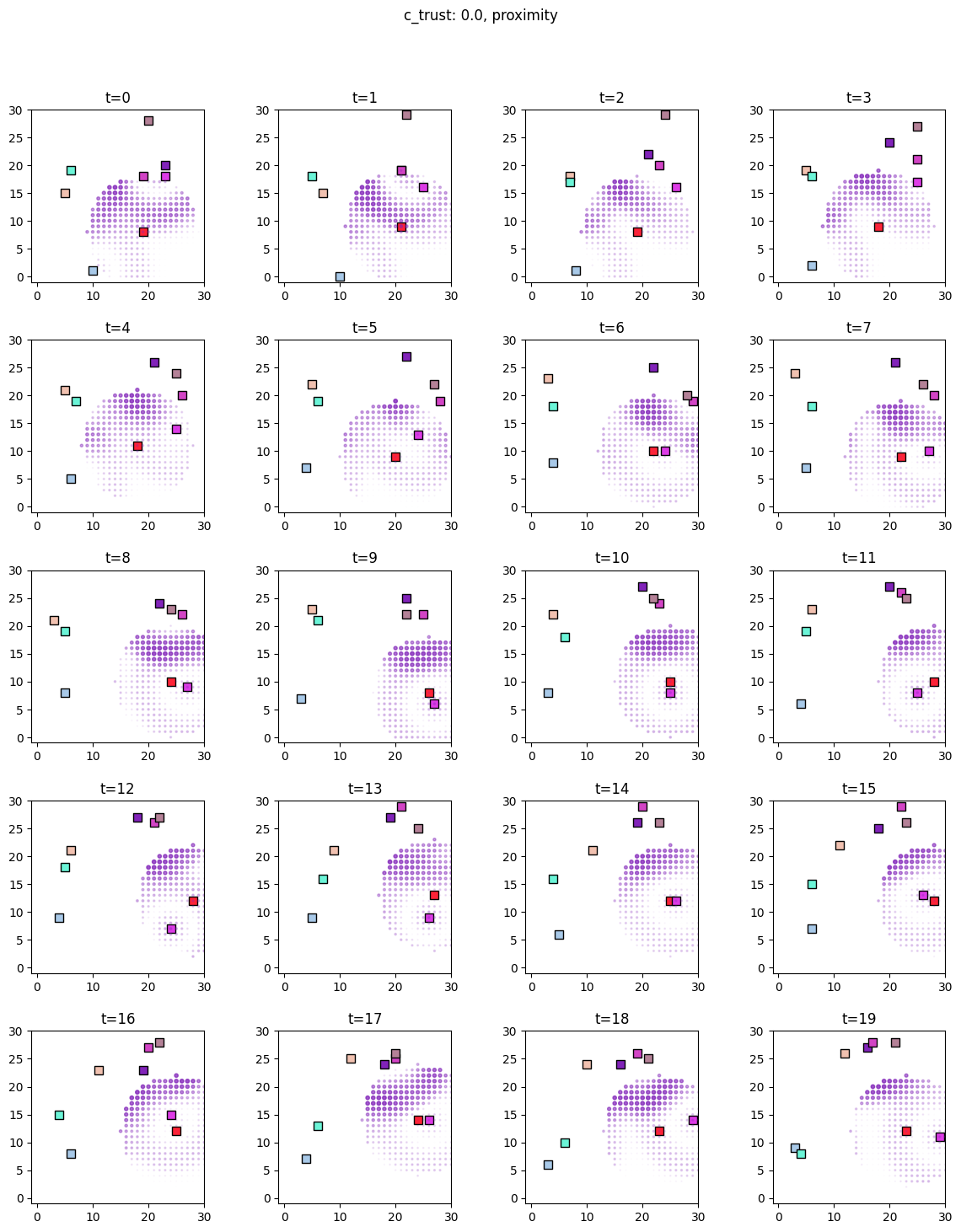
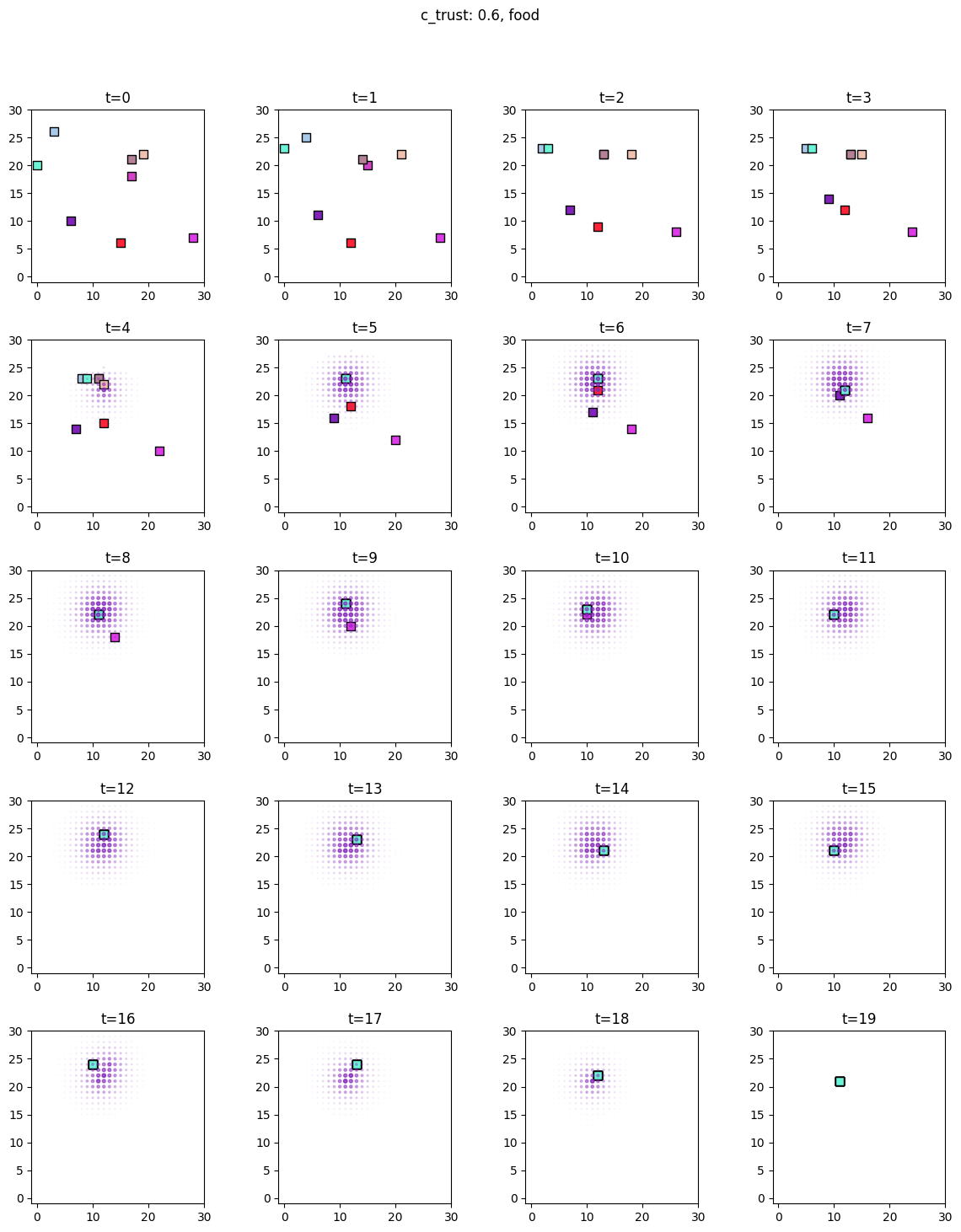
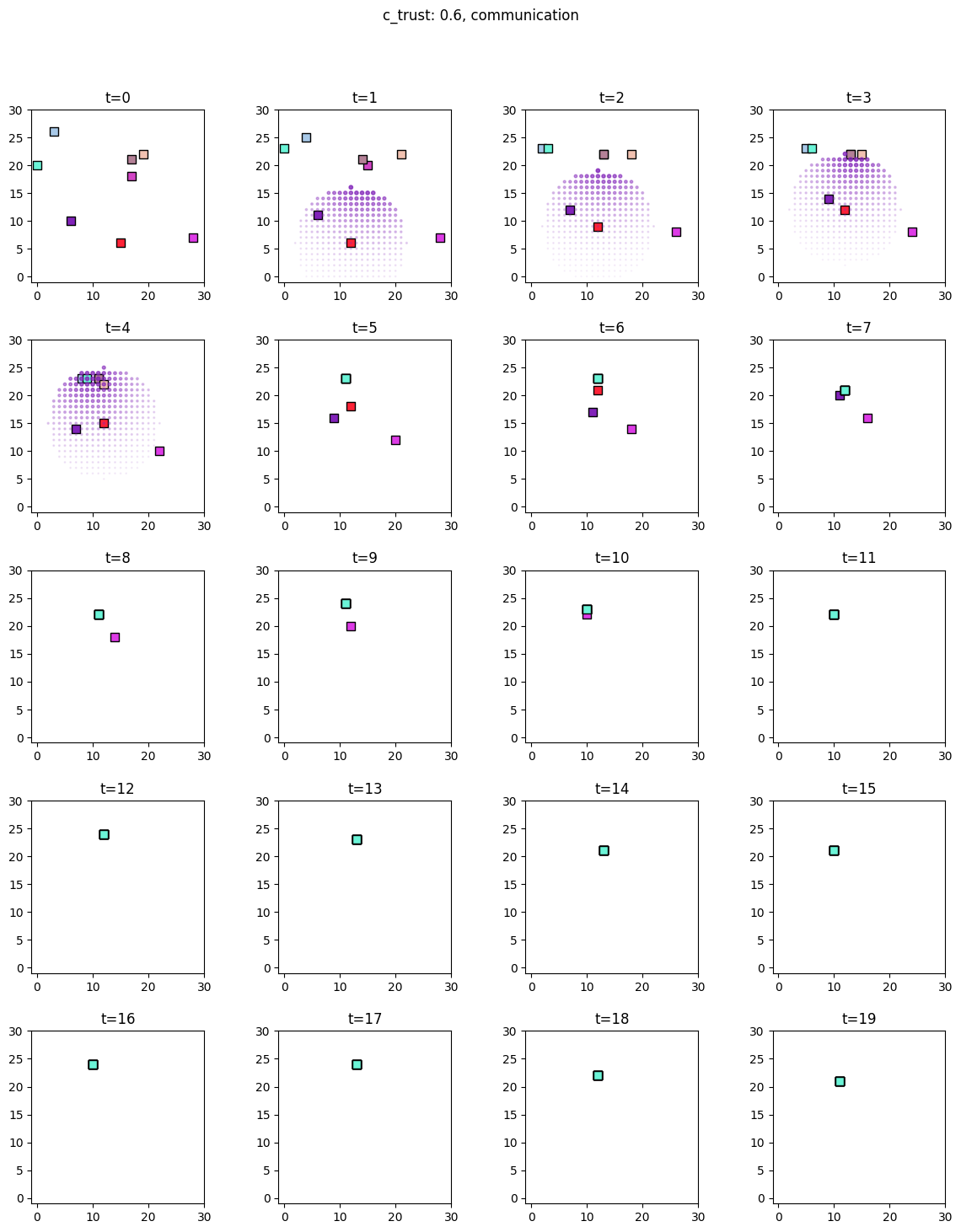
Inference¶
We start by defining the predictors and outcomes for inference.
[8]:
predictors = [
"food_scaled",
"proximity_scaled",
"communication_scaled",
]
outcome_vars = ["nextStep_linear"]
# prepare the training data
predictor_tensors_0, outcome_tensor_0 = ft.prep_data_for_inference(
communicators0, predictors, outcome_vars
)
predictor_tensors_6, outcome_tensor_6 = ft.prep_data_for_inference(
communicators6, predictors, outcome_vars
)
2024-10-31 15:12:02,092: Sample size: 108172
2024-10-31 15:12:02,100: Sample size: 117336
Non-communicating foragers¶
[9]:
# construct Pyro model
model_sigmavar_0 = ft.HeteroskedasticLinear(predictor_tensors_0, outcome_tensor_0)
# runs SVI to approximate the posterior and samples from it
results_0 = ft.get_samples(
model=model_sigmavar_0,
predictors=predictor_tensors_0,
outcome=outcome_tensor_0,
num_svi_iters=1500,
num_samples=1000,
)
selected_sites = [
key
for key in results_0["samples"].keys()
if key.startswith("weight") and not key.endswith("sigma")
]
selected_samples = {key: results_0["samples"][key] for key in selected_sites}
ft.plot_coefs(
selected_samples,
"Non-communicating foragers",
nbins=120,
ann_start_y=160,
ann_break_y=50,
)
2024-10-31 15:12:02,189: Starting SVI inference with 1500 iterations.
[iteration 0001] loss: 427595.0000
[iteration 0200] loss: 310429.3750
[iteration 0400] loss: 313251.0625
[iteration 0600] loss: 310125.8438
[iteration 0800] loss: 309920.7812
[iteration 1000] loss: 309822.6875
[iteration 1200] loss: 309732.7188
[iteration 1400] loss: 309750.7500
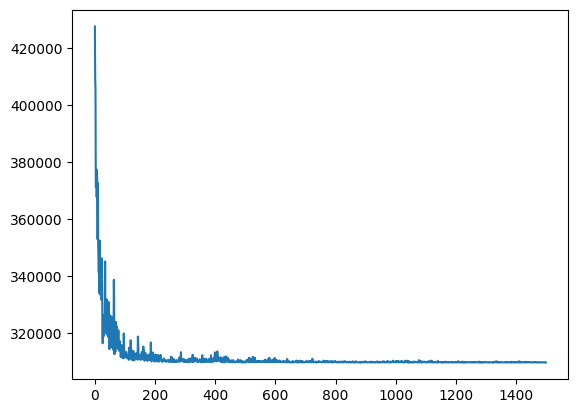
2024-10-31 15:12:20,328: SVI inference completed in 18.14 seconds.
Coefficient marginals:
Site: weight_continuous_food_scaled_nextStep_linear
mean std 5% 25% 50% 75% 95%
0 0.44948 0.007165 0.437664 0.444569 0.449468 0.454571 0.460918
Site: weight_continuous_proximity_scaled_nextStep_linear
mean std 5% 25% 50% 75% 95%
0 -0.125197 0.014528 -0.149991 -0.135048 -0.124864 -0.115152 -0.101205
Site: weight_continuous_communication_scaled_nextStep_linear
mean std 5% 25% 50% 75% 95%
0 -0.068347 0.016553 -0.094638 -0.080031 -0.068555 -0.057606 -0.040602
Communicating foragers¶
[10]:
model_sigmavar_6 = ft.HeteroskedasticLinear(predictor_tensors_6, outcome_tensor_6)
results_6 = ft.get_samples(
model=model_sigmavar_6,
predictors=predictor_tensors_6,
outcome=outcome_tensor_6,
num_svi_iters=1500,
num_samples=1000,
)
selected_sites = [
key
for key in results_6["samples"].keys()
if key.startswith("weight") and not key.endswith("sigma")
]
selected_samples = {key: results_6["samples"][key] for key in selected_sites}
ft.plot_coefs(
selected_samples,
"Communicating foragers",
nbins=120,
ann_start_y=160,
ann_break_y=50,
)
2024-10-31 15:12:21,696: Starting SVI inference with 1500 iterations.
[iteration 0001] loss: 436362.8125
[iteration 0200] loss: 330741.2188
[iteration 0400] loss: 330658.3750
[iteration 0600] loss: 330618.7500
[iteration 0800] loss: 331299.6562
[iteration 1000] loss: 330866.5625
[iteration 1200] loss: 330744.8750
[iteration 1400] loss: 330648.1562
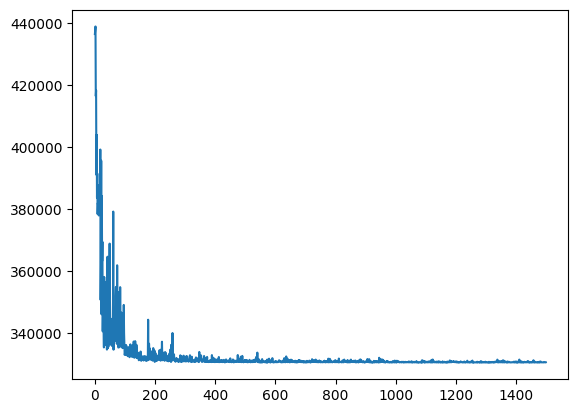
2024-10-31 15:12:40,784: SVI inference completed in 19.09 seconds.
Coefficient marginals:
Site: weight_continuous_food_scaled_nextStep_linear
mean std 5% 25% 50% 75% 95%
0 0.562806 0.007414 0.550546 0.557935 0.562791 0.568074 0.574226
Site: weight_continuous_proximity_scaled_nextStep_linear
mean std 5% 25% 50% 75% 95%
0 0.013317 0.013451 -0.009546 0.00449 0.013319 0.022031 0.035947
Site: weight_continuous_communication_scaled_nextStep_linear
mean std 5% 25% 50% 75% 95%
0 0.143704 0.015194 0.118426 0.133633 0.143564 0.153917 0.170098