Foraging locust with dynamical systems (validation)¶
In this notebook we use the LocustDS class to validate the dynamical systems modeling technique with respect to a training data. Conceptually, we train the same model on different time windows (specified by start time and diffs (duration), both in tens of seconds.) Then, we evaluate them with respect to the same time windows, but using the validation dataset.
The model samples and the results are now saved. The user is welcome to re-run the validation by setting force=True and automatically save the resulting samples by setting save=True in the run.inference() and .validate() methods.
The class that we are using (please go over locust_ds_class.ipynb for a quick tour) is used for each particular validation run. We use the posterior_check() method to evaluate performance of the trained model. For any particular validation run, we pick a time window, train the model on the data from the time window and generate multiple simulations forward form the trained model and take the mean. The distances between the observed counts and the mean simulated
trajectory are the errors (residuals) of the trained model. We use validate() to run analogous calculations for the validation dataset, where the simulated predictions are generated using the same trained model, but the initial state is taken from the validation subset and residuals are calculated between mean predictions for that subset and the actual counts in it. In both cases, the residuals are used to calculate the 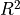 .
.
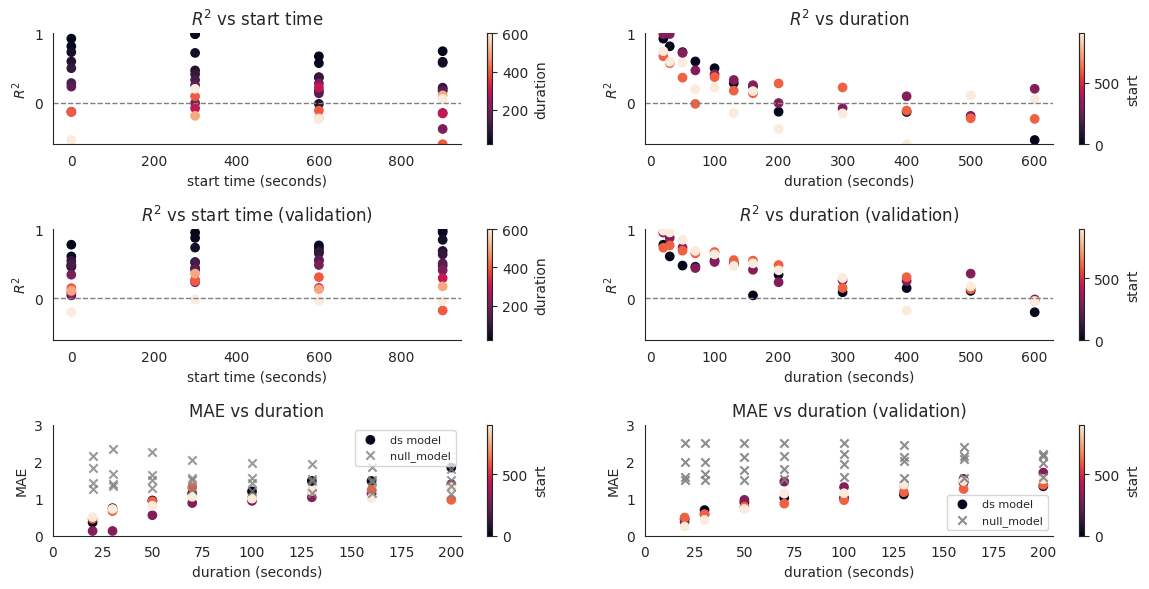
The posterior predictive check and validation results are then plotted against (1) start time, and the intuition is that performance should not differ depending on at which point the time window starts, and (2) duration in seconds, and here it turns out that the dynamical systems display decent performance for predicting the flux of locusts for time windows shorter than 150 seconds, reaching performance comparable with that of the null model for longer periods. As mean absolute errors are easier to interpret, we also plot these against duration both for the training and the validation datasets.
[1]:
import os
import dill
import matplotlib.pyplot as plt
import pandas as pd
import pyro
import seaborn as sns
pyro.settings.set(module_local_params=True)
sns.set_style("white")
# Set seed for reproducibility
seed = 123
pyro.clear_param_store()
pyro.set_rng_seed(seed)
import matplotlib.pyplot as plt
import seaborn as sns
from collab.foraging import locust as lc
from collab.utils import find_repo_root
root = find_repo_root()
smoke_test = "CI" in os.environ
num_iterations = 50 if smoke_test else 1500
num_samples = 20 if smoke_test else 150
starts = [0] if smoke_test else [0, 30, 60, 90]
diffs = [4] if smoke_test else [2, 3, 5, 7, 10, 13, 16, 20, 30, 40, 50, 60]
[2]:
# if the user does not have the samples locally
# the first run will generate
# and save them if save is set to True in appropriate places
data_code = "15EQ20191202"
validation_data_code = "15EQ20191205"
init_ts = []
end_ts = []
null_maes = []
model_maes = []
rsquareds = []
v_null_maes = []
v_model_maes = []
v_rsquareds = []
for start in starts:
for end in [start + diff for diff in diffs]:
init_ts.append(start)
end_ts.append(end)
print(start, end)
locds = lc.LocustDS(
data_code=data_code,
start=start,
end=end,
)
locds.run_inference(
"length",
num_iterations=num_iterations,
num_samples=num_samples,
# force=True, save=True
# uncomment the above to generate the samples anew and save them
)
for key in locds.init_state.keys():
assert (
locds.init_state[key].item() == locds.samples[key][0, 0, 0].item()
), "predictive inits are wrong"
locds.evaluate(
samples=locds.samples,
subset=locds.subset,
figure=False,
# can be used to plot the posterior predictive check
)
null_maes.append(locds.null_mae)
model_maes.append(locds.mae)
rsquareds.append(locds.rsquared)
# uncomment for posterior predicive plots
# if not smoke_test:
# locds.posterior_check()
locds.validate(
validation_data_code=validation_data_code,
# force = True, save=True
figure=False,
)
# same here for validation
for key in locds.v_init_state.keys():
assert (
locds.v_init_state[key].item() == locds.v_samples[key][0, 0, 0].item()
), "validation inits are wrong"
# check if the right initial states are used
# with the right samples
# if you see this error, it's likely that the samples are not saved or loaded correctly
# re-run validation with force=True (and save=True, if you wish to save the samples)
v_null_maes.append(locds.validation[validation_data_code]["null_mae"])
v_model_maes.append(locds.validation[validation_data_code]["mae"])
v_rsquareds.append(locds.validation[validation_data_code]["rsquared"])
# uncomment if you want to plot the posterior validation plots
# one by one
# if not smoke_test:
# locds.posterior_check(
# samples=locds.v_samples,
# subset=locds.v_subset,
# title=f"Validation ({start * 10} to {end * 10})",
# )
0 2
Loading inference samples
Loading validation samples
0 3
Loading inference samples
Loading validation samples
0 5
Loading inference samples
Loading validation samples
0 7
Loading inference samples
Loading validation samples
0 10
Loading inference samples
Loading validation samples
0 13
Loading inference samples
Loading validation samples
0 16
Loading inference samples
Loading validation samples
0 20
Loading inference samples
Loading validation samples
0 30
Loading inference samples
Loading validation samples
0 40
Loading inference samples
Loading validation samples
0 50
Loading inference samples
Loading validation samples
0 60
Loading inference samples
Loading validation samples
30 32
Loading inference samples
Loading validation samples
30 33
Loading inference samples
Loading validation samples
30 35
Loading inference samples
Loading validation samples
30 37
Loading inference samples
Loading validation samples
30 40
Loading inference samples
Loading validation samples
30 43
Loading inference samples
Loading validation samples
30 46
Loading inference samples
Loading validation samples
30 50
Loading inference samples
Loading validation samples
30 60
Loading inference samples
Loading validation samples
30 70
Loading inference samples
Loading validation samples
30 80
Loading inference samples
Loading validation samples
30 90
Loading inference samples
Loading validation samples
60 62
Loading inference samples
Loading validation samples
60 63
Loading inference samples
Loading validation samples
60 65
Loading inference samples
Loading validation samples
60 67
Loading inference samples
Loading validation samples
60 70
Loading inference samples
Loading validation samples
60 73
Loading inference samples
Loading validation samples
60 76
Loading inference samples
Loading validation samples
60 80
Loading inference samples
Loading validation samples
60 90
Loading inference samples
Loading validation samples
60 100
Loading inference samples
Loading validation samples
60 110
Loading inference samples
Loading validation samples
60 120
Loading inference samples
Loading validation samples
90 92
Loading inference samples
Loading validation samples
90 93
Loading inference samples
Loading validation samples
90 95
Loading inference samples
Loading validation samples
90 97
Loading inference samples
Loading validation samples
90 100
Loading inference samples
Loading validation samples
90 103
Loading inference samples
Loading validation samples
90 106
Loading inference samples
Loading validation samples
90 110
Loading inference samples
Loading validation samples
90 120
Loading inference samples
Loading validation samples
90 130
Loading inference samples
Loading validation samples
90 140
Loading inference samples
Loading validation samples
90 150
Loading inference samples
Loading validation samples
[3]:
# put all the predictive check and validation results in data frames
# use date in the file path
# to avoid overwriting previous validation results
# as they require a lot of computation
from datetime import datetime
today = datetime.today().strftime("%Y-%m-%d")
root = find_repo_root()
results_path = os.path.join(
root, f"data/foraging/locust/ds/length_experiment_results_{data_code}_{today}.pkl"
)
v_results_path = os.path.join(
root,
f"data/foraging/locust/ds/length_experiment_v_results_{data_code}_v{validation_data_code}_{today}.pkl",
)
if os.path.exists(results_path):
with open(results_path, "rb") as f:
results = dill.load(f)
else:
results = pd.DataFrame(
{
"start": [_ * 10 for _ in init_ts], # [:-1],
"end": [_ * 10 for _ in end_ts], # [:-1],
"null_mae": null_maes,
"model_mae": model_maes,
"rsquared": rsquareds,
}
)
results["duration"] = [_ * 1 for _ in (results["end"] - results["start"])]
if not smoke_test:
with open(results_path, "wb") as f:
dill.dump(results, f)
if os.path.exists(v_results_path):
with open(v_results_path, "rb") as f:
v_results = dill.load(f)
else:
v_results = pd.DataFrame(
{
"start": [_ * 10 for _ in init_ts],
"end": [_ * 10 for _ in end_ts],
"v_null_mae": v_null_maes,
"v_model_mae": v_model_maes,
"v_rsquared": v_rsquareds,
}
)
v_results["duration"] = [_ * 1 for _ in (v_results["end"] - v_results["start"])]
if not smoke_test:
with open(v_results_path, "wb") as f:
dill.dump(v_results, f)
[4]:
plt.figure(figsize=(12, 6))
plt.subplot(3, 2, 1)
scatter = plt.scatter(
results["start"], results["rsquared"], marker="o", c=results["duration"]
)
plt.axhline(y=0, color="gray", linestyle="--", linewidth=1)
cbar = plt.colorbar(scatter, label="duration")
plt.xlabel("start time (seconds)")
plt.ylabel("$R^2$")
plt.ylim(-0.6, 1)
plt.title("$R^2$ vs start time")
sns.despine()
plt.subplot(3, 2, 2)
scatter2 = plt.scatter(
results["duration"], results["rsquared"], marker="o", c=results["start"]
)
plt.axhline(y=0, color="gray", linestyle="--", linewidth=1)
cbar = plt.colorbar(scatter2, label="start")
plt.xlabel("duration (seconds)")
plt.ylabel("$R^2$")
plt.ylim(-0.6, 1)
plt.title("$R^2$ vs duration")
sns.despine()
plt.subplot(3, 2, 3)
scatter = plt.scatter(
v_results["start"], v_results["v_rsquared"], marker="o", c=v_results["duration"]
)
plt.axhline(y=0, color="gray", linestyle="--", linewidth=1)
cbar = plt.colorbar(scatter, label="duration")
plt.xlabel("start time (seconds)")
plt.ylabel("$R^2$")
plt.ylim(-0.6, 1)
plt.title("$R^2$ vs start time (validation)")
sns.despine()
plt.subplot(3, 2, 4)
scatter2 = plt.scatter(
v_results["duration"], v_results["v_rsquared"], marker="o", c=v_results["start"]
)
plt.axhline(y=0, color="gray", linestyle="--", linewidth=1)
cbar = plt.colorbar(scatter2, label="start")
plt.xlabel("duration (seconds)")
plt.ylabel("$R^2$")
plt.ylim(-0.6, 1)
plt.title("$R^2$ vs duration (validation)")
sns.despine()
plt.subplot(3, 2, 5)
scatter3 = plt.scatter(
results["duration"],
results["model_mae"],
marker="o",
c=results["start"],
label="ds model",
)
plt.scatter(
results["duration"],
results["null_mae"],
marker="x",
c="gray",
alpha=0.8,
label="null_model",
)
# plt.axhline(y=0, color="gray", linestyle="--", linewidth=1)
cbar = plt.colorbar(scatter3, label="start")
plt.xlabel("duration (seconds)")
plt.ylabel("MAE")
plt.ylim(0, 3)
plt.xlim(0, 205)
plt.title("MAE vs duration")
plt.legend(prop={"size": 8})
sns.despine()
plt.subplot(3, 2, 6)
scatter4 = plt.scatter(
v_results["duration"],
v_results["v_model_mae"],
marker="o",
c=v_results["start"],
label="ds model",
)
plt.scatter(
v_results["duration"],
v_results["v_null_mae"],
marker="x",
c="gray",
alpha=0.9,
label="null_model",
)
# plt.axhline(y=0, color="gray", linestyle="--", linewidth=1)
cbar = plt.colorbar(scatter4, label="start")
plt.xlabel("duration (seconds)")
plt.ylabel("MAE")
plt.ylim(0, 3)
plt.xlim(0, 205)
plt.legend(prop={"size": 8})
plt.title("MAE vs duration (validation)")
sns.despine()
plt.tight_layout()
if not smoke_test:
plt.savefig(os.path.join(root, "docs/figures/locust_ds_validation.png"))
plt.show()
[5]:
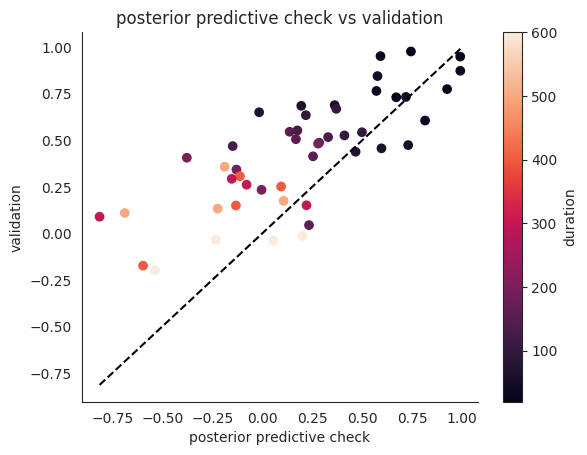
# note validation performance is slightly better, esp. for longer durations
# conjecture: the model slightly overfits early data
# and so loses performance esp. for longer durations
scatter = plt.scatter(
y=v_results["v_rsquared"],
x=results["rsquared"],
marker="o",
c=v_results["duration"],
)
plt.plot(
[results["rsquared"].min(), results["rsquared"].max()],
[results["rsquared"].min(), results["rsquared"].max()],
color="black",
linestyle="--",
)
plt.title("posterior predictive check vs validation")
plt.xlabel("posterior predictive check")
plt.ylabel("validation")
sns.despine()
plt.colorbar(scatter, label="duration")
plt.show()
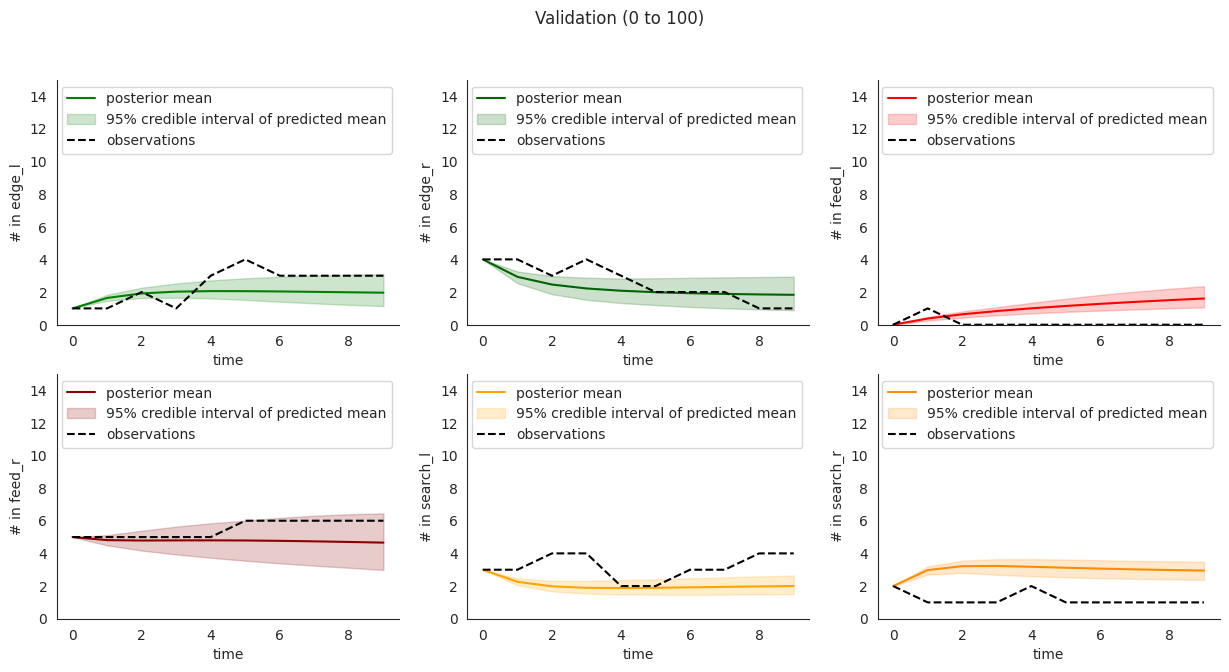
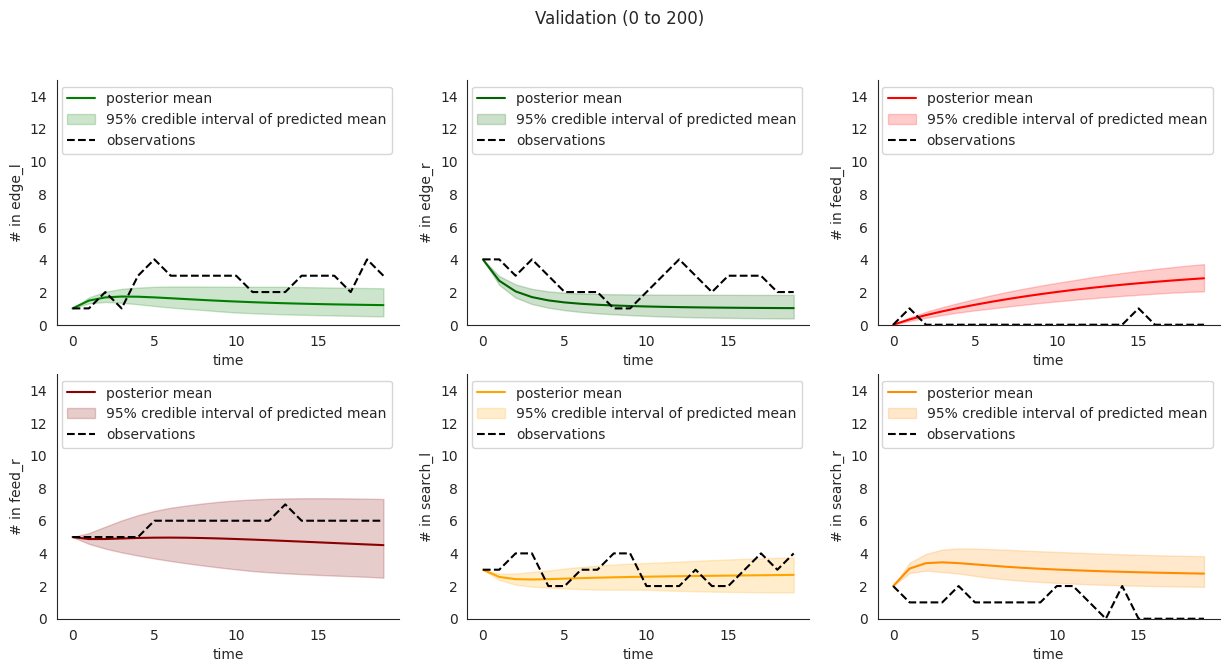
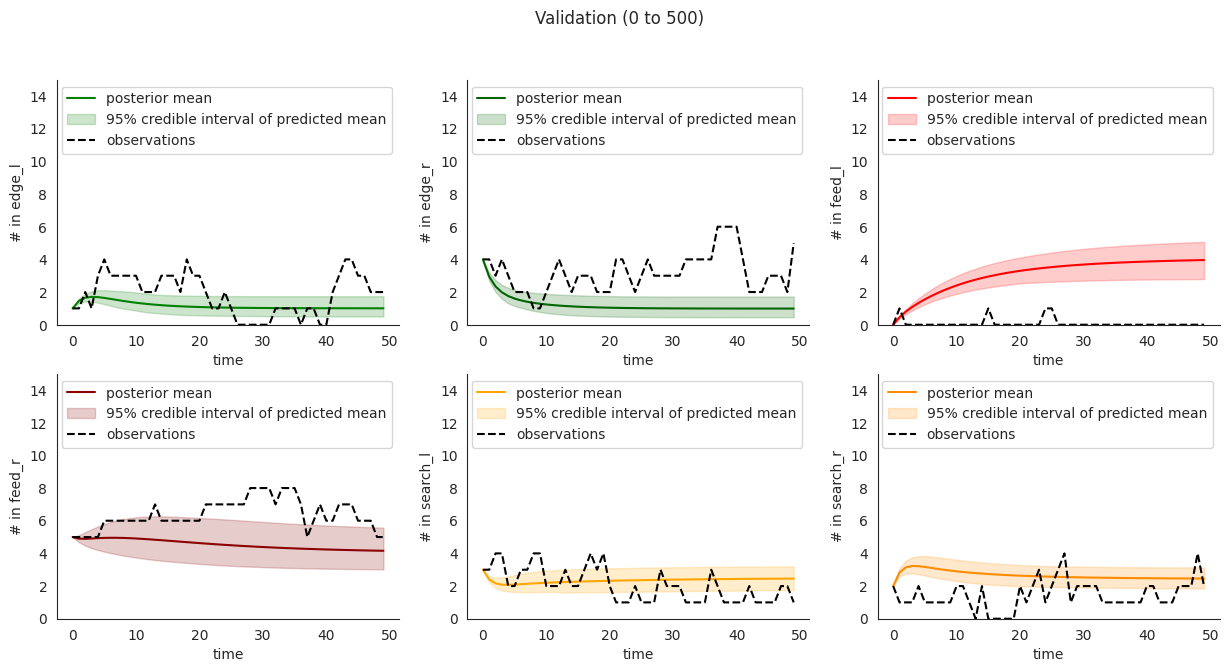
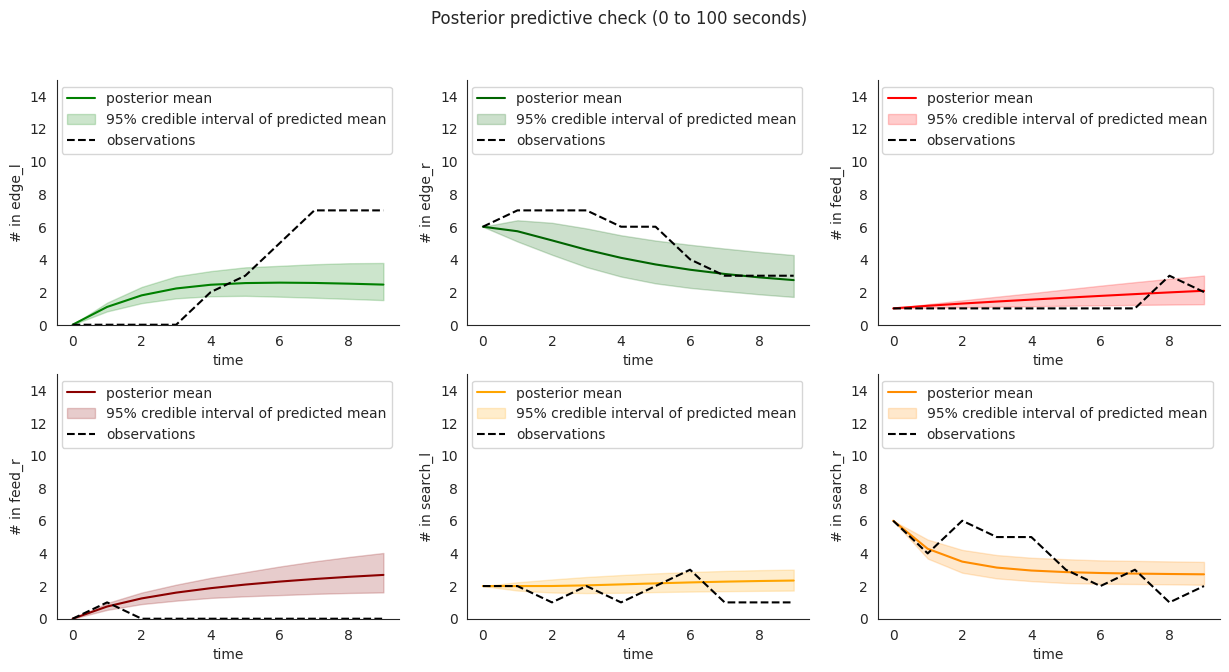
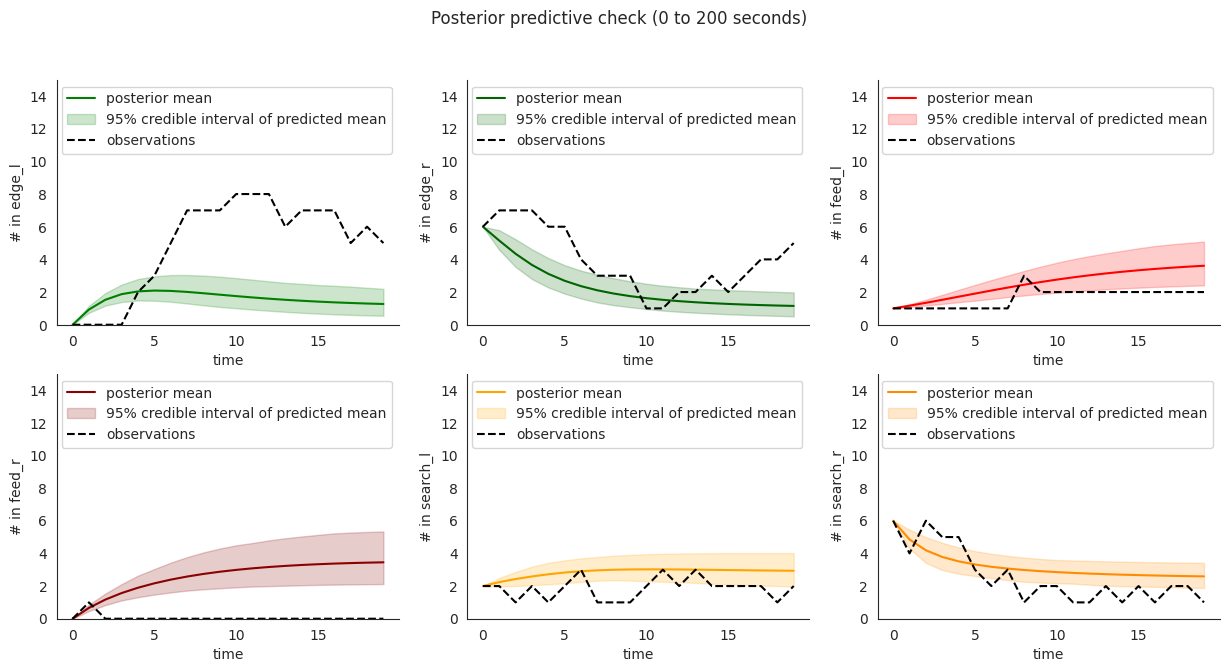
To illustrate, let’s plot posterior checks for a selection of durations. Notice that for longer time windows in some compartments it might happen that the mean predictions might fit the initial few steps and then stabilize at a value that quite far from the overall mean.
[6]:
start = 0
for end in [10, 20, 60]:
locds = lc.LocustDS(
data_code="15EQ20191202",
start=start,
end=end,
)
locds.run_inference(
"length",
num_iterations=num_iterations,
num_samples=num_samples,
lr=0.0005,
# force=True,
save=True,
)
locds.posterior_check()
Loading inference samples
Loading inference samples
Loading inference samples
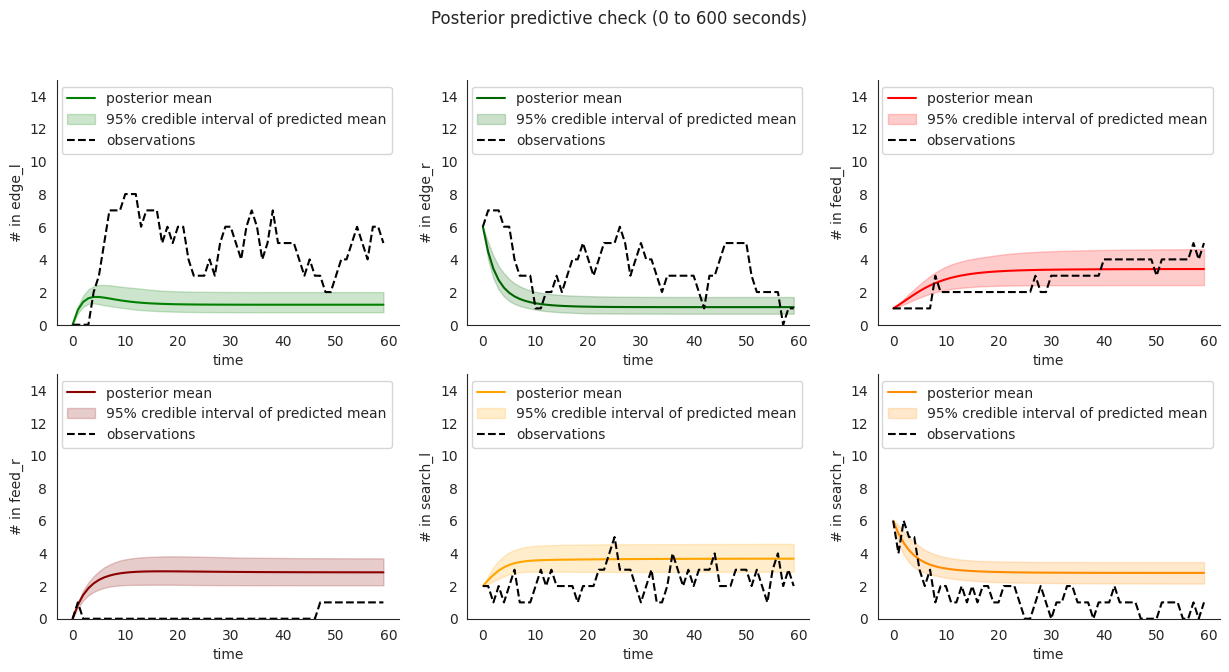
This overfitting to initial steps arises to a lower extent in validation.
[7]:
start = 0
for end in [10, 20, 50]:
locds = lc.LocustDS(
data_code="15EQ20191202",
start=start,
end=end,
)
locds.run_inference(
"length",
num_iterations=num_iterations,
num_samples=num_samples,
lr=0.0005,
# force=True,
save=True,
)
locds.validate(validation_data_code="15EQ20191205", figure=False)
locds.posterior_check(
samples=locds.v_samples,
subset=locds.v_subset,
title=f"Validation ({start * 10} to {end * 10})",
)
Loading inference samples
Loading validation samples
Loading inference samples
Loading validation samples
Loading inference samples
Loading validation samples